
In Stata 16, you can embed and execute Python code from within Stata. Stata's new python command provides a suite of subcommands allowing you to easily call Python from Stata and output Python results within Stata.
You can invoke Python interactively or in do-files and ado-files so that you can leverage Python's extensive language features. You can also execute a Python script file (.py) directly through Stata.
Additionally, the Stata Function Interface (sfi) Python module is included. It provides a bidirectional connection between Stata and Python. It allows you to interact Python's capabilities with Stata's core features. Within the module, classes are defined to provide access to Stata's current dataset, frames, macros, scalars, matrices, value labels, characteristics, global Mata matrices, and more.
Stata supports both Python 2 and Python 3 starting from Python 2.7. You can choose which one to bind to from within Stata.The first time you call python in Stata, Stata will search for Python installations on the system and choose the one with the highest version. Once Stata finds the candidate with the highest version, it will save that information to use in the future. You can then start your Python journey within Stata. Next we will show you how to invoke Python from Stata.
You can type python in the Stata Command Window to enter the Python environment. Think of this as an interactive Python shell. You can use it much like you can use Mata (Stata's built-in matrix programming language) interactively. For example, you could type
. python
| python (type end to exit) | ||
| >>> print('Hello, Python!') | ||
| Hello, Python! | ||
| >>> mylist = ['abcd', 123, 1.23, 'efg'] | ||
| >>> for i in range(3): | ||
| ... print(i) | ||
| ... | ||
| 0 | ||
| 1 | ||
| 2 | ||
| >>> end | ||
| . | ||
It is easy to embed Python code in a do-file. All you need to do is place the Python code within a python and end block.
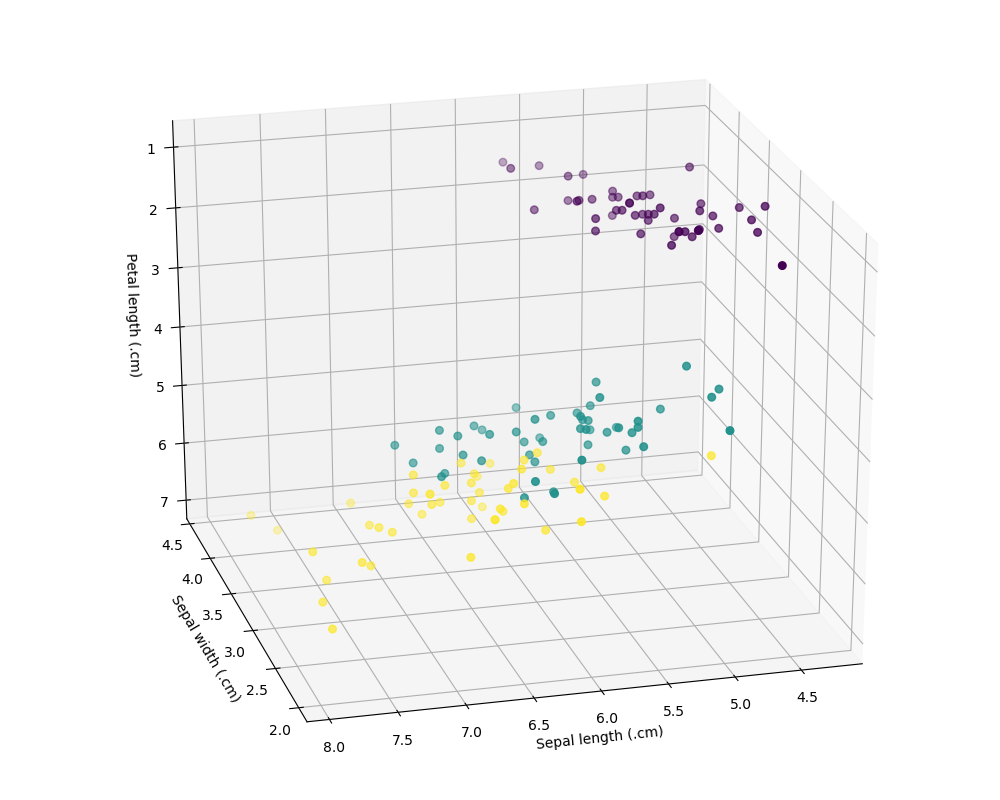
We will use the famous Iris dataset as an illustration. This dataset is used in Fisher's (1936) article. Fisher obtained the Iris data from Anderson (1935). The data consist of four features measured on 50 samples from each of three Iris species. The four features are the length and width of the sepal and petal. The three species are Iris setosa, Iris versicolor, and Iris virginica.
. use http://www.stata-press.com/data/r16/iris, clear
(Iris data)
. describe
Contains data from https://www.stata-press.com/data/r16/iris.dta
obs: 150 Iris data
vars: 6 18 Jan 2018 13:23
(_dta has notes)
| storage display value variable name type format label variable label | ||
| iris byte %10.0g species Iris species seplen double %4.1f Sepal length in cm sepwid double %4.1f Sepal width in cm petlen double %4.1f Petal length in cm petwid double %4.1f Petal width in cm | ||
| Sorted by: Note: Dataset has changed since last saved. | ||
Our goal is to build a classifier using those features to detect the Iris type. Here we will use the Support Vector Machine (SVM) classifier within the scikit-learn Python package to achieve this goal. Note that you need to install the Matplotlib, sklearn, and NumPy packages in your current Python installation to run the following example. Before using Matplotlib with Stata, you may need to set the backend for different Python installations. See Using Matplotlib with Stata for a discussion about calling Matplotlib from Stata. We put the following code in the Do-file Editor and execute it:
use http://www.stata-press.com/data/r16/iris, clear
python:
from sfi import Data
import numpy as np
from sklearn.svm import SVC
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# Use the sfi Data class to pull data from Stata variables into Python
X = np.array(Data.get("seplen sepwid petlen petwid"))
y = np.array(Data.get("iris"))
# Draw a graph in Python and save as samplepy.png
fig = plt.figure(1, figsize=(10, 8))
ax = Axes3D(fig, elev=-155, azim=105)
ax.scatter(X[:, 0], X[:, 1], X[:, 2], c=y, s=30)
ax.set_xlabel("Sepal length (.cm)")
ax.set_ylabel("Sepal width (.cm)")
ax.set_zlabel("Petal length (.cm)")
plt.savefig("samplepy.png")
# Use the data to train C-Support Vector Classifier
svc_clf = SVC(gamma='auto')
svc_clf.fit(X, y)
# Obtain prediction and store back into new Stata variable irispr
irispr = svc_clf.predict(X)
Data.addVarByte('irispr')
Data.store('irispr', None, irispr)
end
* See results in Stata
label values irispr species
label variable irispr predicted
tabulate iris irispr, row
In the code above, we did the following:
We saved the above code in samplepy.do and ran
. do samplepy
which produced the following image and output:
| Key | |||||
| frequency | |||||
| row percentage | |||||
| Iris | predicted | ||||
| species | setosa versicolo virginica | Total | |||
| setosa | 50 0 0 | 50 | |||
| 100.00 0.00 0.00 | 100.00 | ||||
| versicolor | 0 48 2 | 50 | |||
| 0.00 96.00 4.00 | 100.00 | ||||
| virginica | 0 0 50 | 50 | |||
| 0.00 0.00 100.00 | 100.00 | ||||
| Total | 50 48 52 | 150 | |||
| 33.33 32.00 34.67 | 100.00 | ||||
The above table shows that 2 Iris versicolor observations were misclassfied as Iris virginica, and no Iris setosa or Iris virginica were misclassified.
Python code can be embedded and executed in ado-files too. Below, we create a new command mysvm in mysvm.ado to illustrate this purpose. mysvm expects a label variable to be specified first followed by a list of feature variables along with a predict() option specifying the name of the variable where the prediction will be stored.
program mysvm
version 16
syntax varlist, predict(name)
gettoken label feature : varlist
//call the Python function
python: dosvm("`label'", "`feature'", "`predict'")
end
version 16
python:
from sfi import Data
import numpy as np
from sklearn.svm import SVC
def dosvm(label, features, predict):
X = np.array(Data.get(features))
y = np.array(Data.get(label))
svc_clf = SVC(gamma='auto')
svc_clf.fit(X, y)
y_pred = svc_clf.predict(X)
Data.addVarByte(predict)
Data.store(predict, None, y_pred)
end
In the above ado-file, we defined the classifier within the Python function dosvm(), which took the species type variable, the four feature variables, and the new variable storing the predictions as arguments. We called the Python function using the python: istmt syntax in the ado-code.
To produce the same output as above, we can type
. use http://www.stata-press.com/data/r16/iris, clear . describe . mysvm iris seplen sepwid petlen petwid, predict(irispr) . label variable irispr predicted . label values irispr species . tabulate iris irispr, row
Anderson, E. 1935. The irises of the Gaspé Peninsula. Bulletin of the American Iris Society 59: 2–5.
Fisher, R. A. 1936. The use of multiple measurements in taxonomic problems. Annals of Eugenics 7: 179–188.
Hunter, J.D. 2007 "Matplotlib: A 2D Graphics Environment". Computing in Science & Engineering 9: 90–95.
Oliphant, T. E. 2006. A Guide to NumPy, 2nd. Ed. Austin, TX: Continuum Press.
Pedregosa F., G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, E. Duchesnay. 2011. Scikit-learn: Machine Learning in Python. JMLR 12: 2825–2830.
van der Walt, S. C. Colbert, and G. Varoquaux. 2011. The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science & Engineering. 13: 22–30. DOI:10.1109/MCSE.2011.37 (publisher link).
Wikipedia contributors. 2019. Iris flower data set. Wikipedia, The Free Encyclopedia. 2019 Jun 19, 18:30 UTC [cited 2019 Jun 24].
Learn more about Stata's programming features.
Read more about how to call Python from Stata in the Stata Programming Reference Manual; see [P] python.
Read more about the Stata Function Interface (sfi) Python module from stata.com/python.